In the last article, we talked about how to detect missing values. Today we will discuss how to handle the missing values. There are several ways we can use to impute missing values, and the choice of method depends on the nature of data and the problem we are trying to solve.
Let’s start with creating a dataset that contains missing value
import pandas as pd; import numpy as np;
data = {'A':[np.nan,np.nan,1,np.nan,np.nan],
'B':[2,3,4,5,np.nan],
'C':[5,6,7,8,np.nan]}
df =pd.DataFrame(data) 1. Remove missing values
- If there are only small number of missing values (e.g. 3% missing values) and you can afford to lose some rows.
- If there are amounts of number of missing values in some columns (e.g. 95% of missing values)
You can remove the rows or columns with missing values by using dropna()
- Remove the rows where at least one element is missing
- Remove the rows where all elements are missing
- Remove the columns where at least one elements are missing
- Remove the missing elements in a subset scope
# Remove the rows where at least one element is missing
df.dropna()
Out[1]:
A B C
2 1.0 4.0 7.0 # all the missing values are removed
# Remove the rows where all elements are missing
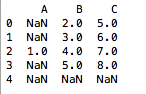
df.dropna(how='all')
Out[2]:
A B C
0 NaN 2.0 5.0
1 NaN 3.0 6.0
2 1.0 4.0 7.0
3 NaN 5.0 8.0 # row 4 is removed
# Remove the columns where at least one elements are missing
df.dropna(axis='columns')
Out[3]:
Empty DataFrame
Columns: []
Index: [0, 1, 2, 3, 4] # because every column contains at least one missing value
# Remove the missing elements in a subset scope
df.dropna(subset=['A','B'])
Out[4]:
A B C
2 1.0 4.0 7.0 # the missing values in the subset are removed2. Impute missing values
If we can not afford the consequence of losing data, we can consider imputing missing values.
2.1 Impute missing values with a Constant
For example, we can fill the missing values with 0.
df.fillna(0) # All the missing values are 0 now
Out[5]:
A B C
0 0.0 2.0 5.0
1 0.0 3.0 6.0
2 1.0 4.0 7.0
3 0.0 5.0 8.0
4 0.0 0.0 0.0We also can fill the missing values with a string.
df.fillna('a') # All the missing values are a now
Out[6]:
A B C
0 a 2 5
1 a 3 6
2 1 4 7
3 a 5 8
4 a a a2.2 Impute missing values with Mean, Median, or Mode
When dealing with continuous numeric data, such as age or income, it’s often appropriate to impute missing values with the mean. Mean imputation assumes that the missing values are missing at random and do not introduce any systematic bias into the data. It helps preserve the overall distribution of the data.
When dealing with skewed data or data with many outliers, you should consider impute missing values with the median. Median imputation can provide a more representative measure of central tendency than the mean.
When dealing with categorical data or discrete data, such as gender, colour, yes or no, you should consider impute missing values with the mode. The mode imputation replaces missing values with the most frequently occurring category in the respective column.
df.fillna(df.mean(), inplace=True) # Fill with column means
df.fillna(df.median(), inplace=True) # Fill with column medians
df.fillna(df.mode().iloc[0], inplace=True) # Fill with column modes2.3 Impute missing values with Forward or Backward Fill
Forward fill and backward fill are handy methods, especially when you’re working with data that follows a sequence, like time-series data. With time-series data, there’s usually a connection between values at different time points. So, if you have gaps in your data, you can use forward fill or backward fill to fill in those missing values by looking at the nearby data points. It’s a practical way to make educated guesses about the missing information in your sequence-based data.
df.fillna(method='ffill', inplace=True) # Forward fill
df.fillna(method='bfill', inplace=True) # Backward fill2.4 Impute missing values with Interpolation
Interpolation is a useful imputation technique in cases where you have missing values in ordered or sequential data, such as time-series data, spatial data, or any data with a natural ordering such as time-series data, E-commerce order data, Sensor data.
df.interpolate(method='linear')
out[]:
A B C
0 NaN 2.0 5.0
1 NaN 3.0 6.0
2 1.0 4.0 7.0
3 1.0 5.0 8.0
4 1.0 5.0 8.0
Further Read:
- ‘linear’: Ignore the index and treat the values as equally spaced. This is the only method supported on MultiIndexes.
- ‘time’: Works on daily and higher resolution data to interpolate given length of interval.
- ‘index’, ‘values’: use the actual numerical values of the index.
- ‘pad’: Fill in NaNs using existing values.
- ‘nearest’, ‘zero’, ‘slinear’, ‘quadratic’, ‘cubic’, ‘barycentric’, ‘polynomial’“
Always evaluate the specific characteristics of your data, the nature of missing, and your domain knowledge before selecting the imputation method, and be aware of its limitations and potential biases.
“
— Ivy Wang
